| 1.A.65 The Coronavirus Viroporin E Protein (Viroporin E) Family
Viroporins are a growing family of viral proteins able to enhance membrane permeability, promoting virus budding. The viroporin activity of the E protein from murine hepatitis virus (MHV), a member of the coronaviruses, resulted in exit of labeled nucleotides from E. coli cells to the cytoplasm upon expression of MHV E. In addition, enhanced entry of the antibiotic hygromycin B occurred at levels comparable to those observed with the viroporin 6K from Sindbis virus. Mammalian cells are also readily permeabilized by the expression of the MHV E protein. Finally, brefeldin A powerfully blocks the viroporin activity of the E protein in BHK cells, suggesting that an intact vesicular system is necessary for this coronavirus to permeabilize mammalian cells (Madan et al., 2005). The E protein has been described as a cation-selective Ca2+ channel (Harrison et al. 2022). The E channel maintains its multi-ionic non-specific neutral character even in concentrated solutions of Ca2+. In contrast to previous studies, Surya et al. 2023 found no evidence that
SARS-CoV-2 E channel activation requires a particular voltage, high
calcium concentrations or low pH, in agreement with available data from
SARS-CoV-1 E. In addition, sedimentation velocity experiments suggest
that the E channel population is mostly pentameric, but very dynamic and
probably heterogeneous, consistent with the broad distribution of
conductance values typically found in electrophysiological experiments.
The latter has been explained by the presence of proteolipidic channel
structures (Surya et al. 2023).
More recently, coronavirus (CoV) envelope (E) protein ion channel activity was determined in channels formed in planar lipid bilayers by peptides representing either the transmembrane domain of severe acute respiratory syndrome CoV (SARS-CoV) E protein, or the full-length E protein. Both of them formed voltage-independent ion-conductive pores with symmetric ion transport properties (Verdiá-Báguena et al., 2012). Mutations N15A and V25F located in the transmembrane domain prevented ion conductivity. E protein derived channels showed no cation preference in non-charged lipid membranes, whereas they behaved as pores with mild cation selectivity in negatively-charged lipid membranes. Thus, the ion conductance was controlled by the lipid composition of the membrane. Lipid charge also regulated the selectivity of a HCoV-229E E protein derived peptide. These results suggested that the lipids are functionally involved in E protein ion channel activity, forming a protein-lipid pore (Verdiá-Báguena et al. 2013). Refinement of the e-protein structure in a native-like environment by molecular dynamics simulations has been achieved, and it shows that it induced local membrane curvature while decreasing local lipid order (Yang et al. 2022). The SARS-CoV-2 envelope protein forms clustered pentamers in lipid bilayers (Somberg et al. 2022).
HydroDock can build hydrated drug-target complexes from scratch. The program requires only the dry target and drug structures and produces their complexes with appropriately positioned water molecules. As a test application of the protocol, Zsidó et al. 2021 built the structures of amantadine derivatives in complex with the influenza M2 transmembrane ion channel. The repositioning of amantadine derivatives from this influenza target to the SARS-CoV-2 envelope protein was also investigated. Excellent agreement was observed between experiments and the structures determined by HydroDock. The atomic resolution complex structures showed that water plays a similar role in the binding of amphipathic amantadine derivatives to transmembrane ion channels of both influenza A and SARS-CoV-2. While the hydrophobic regions of the channels capture the bulky hydrocarbon group of the ligand, the surrounding waters direct its orientation parallel with the axes of the channels via bridging interactions with the ionic ligand head (Zsidó et al. 2021). Mechanisms of host-membrane permeabilization by viroporins has been reviewed (Alcaraz and Nieva 2025).
The generalized reaction catalyzed by the MHV E protein is:
small molecules (out) 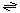 small molecules (in) small molecules (in)
|
This family belongs to the Viroporin-3 Superfamily.
|
| References: |
Alcaraz, A. and J.L. Nieva. (2025). Viroporins: discovery, methods of study, and mechanisms of host-membrane permeabilization. Q. Rev. Biophys. 58: e1.
|
Berta, B., H. Tordai, G.L. Lukács, B. Papp, &.#.1.9.3.;. Enyedi, R. Padányi, and T. Hegedűs. (2024). SARS-CoV-2 envelope protein alters calcium signaling via SERCA interactions. Sci Rep 14: 21200.
|
Breitinger, U., N.K.M. Ali, H. Sticht, and H.G. Breitinger. (2021). Inhibition of SARS CoV Envelope Protein by Flavonoids and Classical Viroporin Inhibitors. Front Microbiol 12: 692423.
|
Cabrera-Garcia, D., R. Bekdash, G.W. Abbott, M. Yazawa, and N.L. Harrison. (2021). The envelope protein of SARS-CoV-2 increases intra-Golgi pH and forms a cation channel that is regulated by pH. J. Physiol. 599: 2851-2868.
|
Cao, Y., R. Yang, W. Wang, I. Lee, R. Zhang, W. Zhang, J. Sun, B. Xu, and X. Meng. (2020). Computational Study of the Ion and Water Permeation and Transport Mechanisms of the SARS-CoV-2 Pentameric E Protein Channel. Front Mol Biosci 7: 565797.
|
Castaño-Rodriguez, C., J.M. Honrubia, J. Gutiérrez-Álvarez, M.L. DeDiego, J.L. Nieto-Torres, J.M. Jimenez-Guardeño, J.A. Regla-Nava, R. Fernandez-Delgado, C. Verdia-Báguena, M. Queralt-Martín, G. Kochan, S. Perlman, V.M. Aguilella, I. Sola, and L. Enjuanes. (2018). Role of Severe Acute Respiratory Syndrome Coronavirus Viroporins E, 3a, and 8a in Replication and Pathogenesis. mBio 9:.
|
Cheng, F.J., C.Y. Ho, T.S. Li, Y. Chen, Y.L. Yeh, Y.L. Wei, T.K. Huynh, B.R. Chen, H.Y. Ko, C.S. Hsueh, M. Tan, Y.C. Wu, H.C. Huang, C.H. Tang, C.H. Chen, C.Y. Tu, and W.C. Huang. (2023). Umbelliferone and eriodictyol suppress the cellular entry of SARS-CoV-2. Cell Biosci 13: 118.
|
Dregni, A.J., M.J. McKay, W. Surya, M. Queralt-Martin, J. Medeiros-Silva, H.K. Wang, V. Aguilella, J. Torres, and M. Hong. (2023). The Cytoplasmic Domain of the SARS-CoV-2 Envelope Protein Assembles into a β-Sheet Bundle in Lipid Bilayers. J. Mol. Biol. 435: 167966.
|
Harrison, N.L., G.W. Abbott, M. Gentzsch, A. Aleksandrov, A. Moroni, G. Thiel, S. Grant, C.G. Nichols, H.A. Lester, A. Hartel, K. Shepard, D.C. Garcia, and M. Yazawa. (2022). How many SARS-CoV-2 "viroporins" are really ion channels? Commun Biol 5: 859.
|
Henke, W., H. Waisner, S.P. Arachchige, M. Kalamvoki, and E. Stephens. (2022). The Envelope Proteins from SARS-CoV-2 and SARS-CoV Potently Reduce the Infectivity of Human Immunodeficiency Virus type 1 (HIV-1). Res Sq.
|
Hong, M., V. Mandala, M. McKay, A. Shcherbakov, A. Dregni, and A. Kolocouris. (2020). Structure and Drug Binding of the SARS-CoV-2 Envelope Protein in Phospholipid Bilayers. Res Sq.
|
Jalily, P.H., H. Jalily Hasani, and D. Fedida. (2022). In Silico Evaluation of Hexamethylene Amiloride Derivatives as Potential Luminal Inhibitors of SARS-CoV-2 E Protein. Int J Mol Sci 23:.
|
Lu, H., Z. Liu, X. Deng, S. Chen, R. Zhou, R. Zhao, R. Parandaman, A. Thind, J. Henley, L. Tian, J. Yu, L. Comai, P. Feng, and W. Yuan. (2023). Potent NKT cell ligands overcome SARS-CoV-2 immune evasion to mitigate viral pathogenesis in mouse models. PLoS Pathog 19: e1011240.
|
Madan, V., J. García Mde, M.A. Sanz, and L. Carrasco. (2005). Viroporin activity of murine hepatitis virus E protein. FEBS Lett. 579(17):3607-3612.
|
Mandala, V.S., M.J. McKay, A.A. Shcherbakov, A.J. Dregni, A. Kolocouris, and M. Hong. (2020). Structure and drug binding of the SARS-CoV-2 envelope protein transmembrane domain in lipid bilayers. Nat Struct Mol Biol 27: 1202-1208.
|
Medeiros-Silva, J., A.J. Dregni, N.H. Somberg, P. Duan, and M. Hong. (2023). Atomic structure of the open SARS-CoV-2 E viroporin. Sci Adv 9: eadi9007.
|
Medeiros-Silva, J., N.H. Somberg, H.K. Wang, M.J. McKay, V.S. Mandala, A.J. Dregni, and M. Hong. (2022). pH- and Calcium-Dependent Aromatic Network in the SARS-CoV-2 Envelope Protein. J. Am. Chem. Soc. 144: 6839-6850.
|
Nieto-Torres JL., Verdia-Baguena C., Jimenez-Guardeno JM., Regla-Nava JA., Castano-Rodriguez C., Fernandez-Delgado R., Torres J., Aguilella VM. and Enjuanes L. (2015). Severe acute respiratory syndrome coronavirus E protein transports calcium ions and activates the NLRP3 inflammasome. Virology. 485:330-9.
|
Park, S.H., H. Siddiqi, D.V. Castro, A.A. De Angelis, A.L. Oom, C.A. Stoneham, M.K. Lewinski, A.E. Clark, B.A. Croker, A.F. Carlin, J. Guatelli, and S.J. Opella. (2021). Interactions of SARS-CoV-2 envelope protein with amilorides correlate with antiviral activity. PLoS Pathog 17: e1009519.
|
Ruch, T.R. and C.E. Machamer. (2012). A single polar residue and distinct membrane topologies impact the function of the infectious bronchitis coronavirus e protein. PLoS Pathog 8: e1002674.
|
Scott, C. and S. Griffin. (2015). Viroporins: structure, function and potential as antiviral targets. J Gen Virol 96: 2000-2027.
|
Somberg, N.H., J. Medeiros-Silva, H. Jo, J. Wang, W.F. DeGrado, and M. Hong. (2023). Hexamethylene Amiloride Binds the SARS-CoV-2 Envelope Protein at the Protein-Lipid Interface. Protein. Sci. e4755. [Epub: Ahead of Print]
|
Somberg, N.H., W.W. Wu, J. Medeiros-Silva, A.J. Dregni, H. Jo, W.F. DeGrado, and M. Hong. (2022). SARS-CoV-2 Envelope Protein Forms Clustered Pentamers in Lipid Bilayers. Biochemistry 61: 2280-2294.
|
Surya, W., E. Tavares-Neto, A. Sanchis, M. Queralt-Martín, A. Alcaraz, J. Torres, and V.M. Aguilella. (2023). The Complex Proteolipidic Behavior of the SARS-CoV-2 Envelope Protein Channel: Weak Selectivity and Heterogeneous Oligomerization. Int J Mol Sci 24:.
|
Surya, W., Y. Li, C. Verdià-Bàguena, V.M. Aguilella, and J. Torres. (2015). MERS coronavirus envelope protein has a single transmembrane domain that forms pentameric ion channels. Virus Res 201: 61-66.
|
Takano T., Nakano K., Doki T. and Hohdatsu T. (2015). Differential effects of viroporin inhibitors against feline infectious peritonitis virus serotypes I and II. Arch Virol. 160(5):1163-70.
|
To, J., W. Surya, T.S. Fung, Y. Li, C. Verdià-Bàguena, M. Queralt-Martin, V.M. Aguilella, D.X. Liu, and J. Torres. (2016). Channel inactivating mutations and their revertant mutants in the envelope protein of the infectious bronchitis virus. J. Virol. [Epub: Ahead of Print]
|
Torres, J., K. Parthasarathy, X. Lin, R. Saravanan, A. Kukol, and D.X. Liu. (2006). Model of a putative pore: the pentameric α-helical bundle of SARS coronavirus E protein in lipid bilayers. Biophys. J. 91: 938-947.
|
Torres, J., U. Maheswari, K. Parthasarathy, L. Ng, D.X. Liu, and X. Gong. (2007). Conductance and amantadine binding of a pore formed by a lysine-flanked transmembrane domain of SARS coronavirus envelope protein. Protein. Sci. 16: 2065-2071.
|
Townsend, J.A., O. Fapohunda, Z. Wang, H. Pham, M.T. Taylor, B. Kloss, S.H. Park, S. Opella, C.A. Aspinwall, and M.T. Marty. (2024). Differences in Oligomerization of the SARS-CoV-2 Envelope Protein, Poliovirus VP4, and HIV Vpu. Biochemistry. [Epub: Ahead of Print]
|
Verdia-Baguena C., Nieto-Torres JL., Alcaraz A., Dediego ML., Enjuanes L. and Aguilella VM. (2013). Analysis of SARS-CoV E protein ion channel activity by tuning the protein and lipid charge. Biochim Biophys Acta. 1828(9):2026-31.
|
Verdiá-Báguena, C., J.L. Nieto-Torres, A. Alcaraz, M.L. Dediego, J. Torres, V.M. Aguilella, and L. Enjuanes. (2012). Coronavirus E protein forms ion channels with functionally and structurally-involved membrane lipids. Virology 432: 485-494.
|
Wang, C.W. and W.B. Fischer. (2022). Rotational Dynamics of The Transmembrane Domains Play an Important Role in Peptide Dynamics of Viral Fusion and Ion Channel Forming Proteins-A Molecular Dynamics Simulation Study. Viruses 14:.
|
Yadav, R., C. Choudhury, Y. Kumar, and A. Bhatia. (2022). Virtual repurposing of ursodeoxycholate and chenodeoxycholate as lead candidates against SARS-Cov2-Envelope protein: A molecular dynamics investigation. J Biomol Struct Dyn 40: 5147-5158.
|
Yang, R., S. Wu, S. Wang, G. Rubino, J.D. Nickels, and X. Cheng. (2022). Refinement of SARS-CoV-2 envelope protein structure in a native-like environment by molecular dynamics simulations. Front Mol Biosci 9: 1027223.
|
Zhang, R., H. Qin, R. Prasad, R. Fu, H.X. Zhou, and T.A. Cross. (2023). Dimeric Transmembrane Structure of the SARS-CoV-2 E Protein. bioRxiv.
|
Zsidó, B.Z., R. Börzsei, V. Szél, and C. Hetényi. (2021). Determination of Ligand Binding Modes in Hydrated Viral Ion Channels to Foster Drug Design and Repositioning. J Chem Inf Model 61: 4011-4022.
|
| Examples: |
| TC# | Name | Organismal Type | Example |
| 1.A.65.1.1 | The envelope (E) viroporin protein of 85 aas and 2 TMSs. | Virus | E protein of Murine Hepatitis Virus (MHV) (83aas; P0C2R0) |
| |
| 1.A.65.1.2 | The SARS coronavirus pore-forming envelope (E) protein or protein 3a (76 aas; 1 TMS) forms a pentameric cation-selective pore (Torres et al. 2006; Scott and Griffin 2015) that binds amantadine (Torres et al., 2007). A single polar residue and distinct membrane topologies impact its function (Ruch and Machamer, 2012). The E protein ion channel (IC) activity is cation-specific and K+-selective and is specifically correlated with enhanced pulmonary damage, edema accumulation and death. Calcium ions together with pH modulated E protein pore charge and selectivity (Nieto-Torres et al. 2015). There is a single transmembrane domain in E, suggesting an allosteric
interaction between extramembrane and transmembrane domains (To et al. 2016). | Virus | Protein E of SARS (NP_828854) (Q19QW7) |
| |
| 1.A.65.1.3 | Envelope small membrane viroporin protein of 82 aas and 1 or 2 TMSs, protein E or sM. Viroporin inhibitors have been identified (Takano et al. 2015). | Viruses | Viroporin of feline infectious peritonitis virus (FIPV) |
| |
| 1.A.65.1.4 | MERS CoV Viroporin of 82 aas and 1 TMS. Induces the formation of pentameric hydrophilic pores in cellular membranes followed by apoptosis (Surya et al. 2015). | Viruses | Viroporin of Human Middle East respiratory syndrome coronavirus (MERS CoV) or
EMC (HCoV-EMC) |
| |
| 1.A.65.1.5 | ORF5-E fusion protein of 194 aa | | Orf5-E of Middle East respiratory syndrome-related coronavirus |
| |
| 1.A.65.1.6 | Envelope protein of 75 aas and 1 TMS. | | Envelope small protein of Alphacoronavirus Bat-CoV/P. kuhlii |
| |
| 1.A.65.1.7 | Envelope (E) viroporin protein, ORF5, of 75 aas and 1 N-terminal TMS. The E-proteins of CoV, CoV-2 and MERS oligomerize to form homopentamers by aligning their TMSs into a pore-forming complex in phospholipid membranes (Surya et al. 2015). The pore is weakly cation selective with Ca2+ favored over K+, and Na+ favored over H+ (Castaño-Rodriguez et al. 2018). It is involved in various aspects of the virus life cycle including assembly, budding, envelope formation, virus release, and inflammasome activation (Breitinger et al. 2021). The structure and drug binding of the SARS-CoV-2 Envelope (E) protein in phospholipid bilayers has been determined (Hong et al. 2020). E forms a five-helix bundle surrounding a narrow central pore. The middle of the TM segment is distorted from the ideal α-helical geometry due to three regularly spaced phenylalanine residues, which stack within each helix and between neighboring helices. These aromatic interactions, together with interhelical Val and Leu interdigitation, cause a dehydrated pore compared to the viroporins of influenza and HIV viruses. Hexamethylene amiloride and amantadine bind shallowly to polar residues at the N-terminal lumen, while acidic pH affects the C-terminal conformation. Thus, SARS-CoV-2 E forms a structurally robust but bipartite channel whose N- and C-terminal halves can interact with drugs, ions and other viral and host proteins semi-independently (Hong et al. 2020). Mandala et al. 2020 reported a 2.1-Å structure and the drug-binding site of E's transmembrane domain (ETM), determined using solid-state NMR spectroscopy. In lipid bilayers that mimic the endoplasmic reticulum-Golgi intermediate compartment (ERGIC) membrane, ETM forms a five-helix bundle surrounding a narrow pore. The protein deviates from the ideal alpha-helical geometry due to three phenylalanine residues, which stack within each helix and between helices. Together with valine and leucine interdigitation, these cause a dehydrated pore compared with the viroporins of influenza viruses and HIV. Hexamethylene amiloride binds the polar amino-terminal lumen, whereas acidic pH affects the carboxy-terminal conformation. Thus, the N- and C-terminal halves of this bipartite channel may interact with other viral and host proteins semi-independently. The structure sets the stage for designing E inhibitors as antiviral drugs (Mandala et al. 2020). Chenodeoxycholate(CDC) and ursodeoxycholate (UDC) bind to the envelope (E) protein of SARS-Cov2 and serve as candidates to hinder the survival of SARS-Cov2 by disrupting the structure of SARS-Cov2-E and facilitating the entry of solvents/polar inhibitors inside the viral cell (Yadav et al. 2022). Interactions of SARS-CoV-2 envelope protein with amilorides promote antiviral activity (Park et al. 2021). E-protein mediated currents were inhibited by amantadine and rimantadine, as well as 5-(N,N-hexamethylene)amiloride (HMA). Of 10 flavonoids, epigallocatechin and quercetin were most effective (Breitinger et al. 2021). The e-protein increases the intra-Golgi pH by forming a cation channel that is regulated by pH(Cabrera-Garcia et al. 2021). A cell-based system combined with flow cytometry has been used to evaluate
antibody responses against SARS-CoV-2 transmembrane proteins in
patients with COVID-19 (Martin et al. 2022). An intricate aromatic network regulates the opening of the ETM channel pore (Medeiros-Silva et al. 2022). Rotational dynamics of the transmembrane domains play important roles in peptide dynamics of viral fusion and ion channel forming proteins (Wang and Fischer 2022). Hexamethylene amiloride derivatives are potential luminal inhibitors of the SARS-CoV-2 E Protein (Jalily et al. 2022). The envelope proteins from SARS-CoV-2 and SARS-CoV potently reduce the infectivity of human immunodeficiency virus type 1 (HIV-1) (Henke et al. 2022). The cytoplasmic domain of the SARS-CoV-2 envelope protein assembles into a beta-sheet bundle in lipid bilayers (Dregni et al. 2023). The E protein of SARS-CoV-2 efficiently down-regulates the cell surface expression of the antigen-presenting molecule, CD1d, to suppress the function of iNKT cells. E protein plays roles in virion packaging and envelopment during viral morphogenesis. The transmembrane domain of E protein is responsible for suppressing CD1d expression by specifically reducing the level of mature, post-ER forms of CD1d, suggesting that it suppressed the trafficking of CD1d proteins and leads to their degradation. Point mutations demonstrated that the putative ion channel function is required for suppression of CD1d expression, and inhibition of the ion channel function using small chemicals rescued CD1d expression (Lu et al. 2023). However, Zhang et al. 2023 identified a symmetric helix-helix interface, leading to the prediction of a dimeric structure that does not support channel activity. The two helices have a tilt angle of only 6 degrees , resulting in an extended interface dominated by Leu and Val side chains. While residues Val14-Thr35 are almost all buried in the hydrophobic region of the membrane, Asn15 lines a water-filled pocket that potentially serves as a drug-binding site. The E and other viral proteins may adopt different oligomeric states to help perform multiple functions (Zhang et al. 2023). Umbelliferone and eriodictyol suppress the cellular entry of SARS-CoV-2 (Cheng et al. 2023). Hexamethylene amiloride binds the SARS-CoV-2 Envelope (E) protein at the protein-lipid interface (Somberg et al. 2023). The atomic structure of the open SARS-CoV-2 E viroporin has been determined (Medeiros-Silva et al. 2023). This E protein is a specific dimer, VP4 of poliovirus is exclusively monomeric, and Vpu
of HIV assembles into a polydisperse mixture of oligomers under these
conditions. Overall, these results revealed the diversity in the
oligomerization of viroporins (Townsend et al. 2024). The E protein perturbes Ca2+ homeostasis. It is structurally similar to regulins such as phospholamban, that regulate the sarco/endoplasmic
reticulum calcium ATPases (SERCA). The
SARS-CoV-2 E protein affects SERCA as an exoregulin and forms oligomers with
regulins, and thus alters the monomer/multimer regulin ratio thereby influencing their interactions with SERCAs. A direct interaction between E protein and SERCA2b results in a
decrease in SERCA-mediated ER Ca2+ reload (Berta et al. 2024). | | E-protein of severe acute respiratory syndrome coronavirus 2, SARS-CoV-2 |
| |
| 1.A.65.1.8 | Protein-E of 78 aas and 2 TMSs. | | E-protein of rodent coronavirus |
| |
| 1.A.65.1.9 | E-protein of 89 aas and 2 TMSs | | E-protein of rabbit coronavirus |
| |
 small molecules (in)
small molecules (in)