| 3.B.1 The Na+-transporting Carboxylic Acid Decarboxylase (NaT-DC) Family
Porters of the NaT-DC family catalyze decarboxylation of a substrate carboxylic acid and use the energy released to drive extrusion of one or two sodium ions (Na+) from the cytoplasm of the cell (Boiangiu et al., 2005). These systems have been characterized only from bacteria. Distinct enzymes catalyze decarboxylation of (1) oxaloacetate, (2) methylmalonyl-CoA, (3) glutaconyl-CoA and (4) malonate. The oxaloacetate decarboxylases (EC 4.1.1.3), methylmalonyl CoA decarboxylases (EC 4.1.1.4) and malonate decarboxylases are homologous. Glutaconyl-CoA decarboxylase (EC 4.1.1.70) consists of four subunits: α (GcdA, 587 aas; catalytic subunit), β (GcdB, 375 aas; 9 TMSs; Na+-transporter subunit), γ (GcdC, 145 aas; biotin-carrier subunit) and δ (GcdD, 107 aas; 1 TMS; the GcdA anchor protein).
All four enzyme porters are biotin-containing multisubunit enzymes. The α-δ subunits of these enzymes are homologous to proteins encoded within the genomes of archaea, such as Pyrococcus abyssi (Cohen et al., 2003). Consequently, NaT-DC family members may be present in archaea as well as bacteria.
The α-subunits of the oxaloacetate and methylmalonyl-CoA decarboxylases are homologous to many biotin-containing enzymes including (1) pyruvate carboxylases, (2) homocitrate synthases, (3) biotin carboxyl carrier proteins, (4) isopropylmalate synthases and (5) acyl-CoA carboxylase. The α-subunit of the glutaconate decarboxylase is homologous to propionyl-CoA carboxylase. The crystal structure of the carboxyltransferase at 1.7 A resolution shows a
dimer of alpha(8)beta(8) barrels with an active site metal ion,
identified spectroscopically as Zn2+ (Granjon et al. 2010).
The high resolution crystal structure of the α-subunit of the glutaconyl CoA decarboxylase (Gcdα) of Acidaminococcus fermentans (TC #3.B.1.1.3) has been solved (Wendt et al., 2003). The active site of the dimeric enzyme lies at the interface between the two monomers. The N-terminal domain binds the glutaconyl-CoA, and the C-terminal domain binds the biotinyl lysine moiety. The enzyme transfers CO2 from glutaconyl-CoA to a biotin carrier protein (the γ-subunit) that is subsequently decarboxylated by the carboxybiotin decarboxylation site within the Na+ pumping beta subunit (Gcdβ). A proposed structure of the holoenzyme positions the water-filled central channel of the Gcdα dimer coaxial with the ion channel in Gcdβ. The central channel is blocked by arginines which could allow Na+ passage by conformational movement or by entry through two side channels (Wendt et al., 2003).
The β-subunits possess 9 transmembrane α-helical spanners (TMSs), and the protein may dip into the membrane twice between TMSs III and IV. The most conserved regions are segments IIIa (the first membrane loop following TMS III) and TMS VIII. Conserved residues therein, D203 (IIIa), Y229 (IV) and N373, G377, S382 and R389 (VIII), provide Na+ binding sites and the translocation pathway. D203 and S382 may provide two binding sites for the two Na+ ions. D203 is absolutely essential for function and may provide the primary intramembranous Na+-binding site. The beta subunits of these transporters show sufficient sequence similarity to the Na+:H+ antiporters of the CPA2 family (TC #2.A.37) to establish homology (K. Studley and M.H. Saier, Jr., unpublished results).
The generalized reaction for the NaT-DC family is:
R - CO2- H+ (out) 1 or 2 Na+ (in) 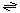 R - H CO2 1 or 2 Na+ (out) R - H CO2 1 or 2 Na+ (out)
|
This family belongs to the CPA Superfamily.
|
| References: |
Balsera, M., R.M. Buey, and X.D. Li. (2011). Quaternary structure of the oxaloacetate decarboxylase membrane complex and mechanistic relationships to pyruvate carboxylases. J. Biol. Chem. 286: 9457-9467.
|
Berg, M., H. Hilbi, and P. Dimroth. (1997). Sequence of a gene cluster from Malonomonas rubra encoding components of the malonate decarboxylase Na+ pump and evidence for their function. Eur. J. Biochem. 245: 103-105.
|
Boiangiu, C.D., E. Jayamani, D. Brugel, G. Herrmann, J. Kim, L. Forzi, R. Hedderich, I. Vgenopoulou, A.J. Pierik, J. Steuber, and W. Buckel. (2005). Sodium ion pumps and hydrogen production in glutamate fermenting anaerobic bacteria. J. Mol. Microbiol. Biotechnol. 10: 105-119.
|
Braune, A., K. Bendrat, S. Rospert, and W. Buckel. (1999). The sodium ion translocating glutaconyl-CoA decarboxylase from Acidaminococcus fermentans: cloning and function of the genes forming a second operon. Mol. Microbiol. 31: 473-487.
|
Buckel, W. (2001). Sodium ion-translocating decarboxylases. Biochim. Biophys. Acta 1505: 15-27.
|
Cohen, G.N., V. Barbe, D. Flament, M. Galperin, R. Heilig, O. Lecompte, O. Poch, D. Prieur, J. Quérellou, R. Ripp, J.-C. Thierry, J. Van der Oost, J. Weissenbach, Y. Zivanovic, and P. Forterre. (2003). An integrated analysis of the genome of the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol. 47: 1495-1512.
|
Di Bernardino, M. and P. Dimroth. (1996). Aspartate 203 of the oxaloacetate decarboxylase β-subunit catalyses both the chemical and vectorial reaction of the Na+ pump. EMBO J. 15: 1842-1849.
|
Dimroth, P. (1997). Primary sodium ion translocating enzymes. Biochim. Biophys. Acta 1318: 11-51.
|
Dimroth, P. and B. Schink. (1998). Energy conservation in the decarboxylation of dicarboxylic acids by fermenting bacteria. Arch. Microbiol. 170: 69-77.
|
Dimroth, P., P. Jockel, and M. Schmid. (2001). Coupling mechanism of the oxaloacetate decarboxylase Na+ pump. Biochim. Biophys. Acta 1505: 1-14.
|
Granjon, T., O. Maniti, Y. Auchli, P. Dahinden, R. Buchet, O. Marcillat, and P. Dimroth. (2010). Structure-function relations in oxaloacetate decarboxylase complex. Fluorescence and infrared approaches to monitor oxomalonate and Na+ binding effect. PLoS One 5: e10935.
|
Huder, J.B. and P. Dimroth. (1993). Sequence of the sodium ion pump methylmalonyl-CoA decarboxylase from Veillonella parvula. J. Biol. Chem. 268: 24564-24571.
|
Huder, J.B. and P. Dimroth. (1995). Expression of the sodium ion pump methylmalonyl-coenzyme A-decarboxylase from Veillonella parvula and of mutated enzyme specimens in Escherichia coli. J. Bacteriol. 177: 3623-3630.
|
Inoue, M. and X. Li. (2015). Highly active and stable oxaloacetate decarboxylase Na⁺ pump complex for structural analysis. Protein Expr Purif 115: 34-38.
|
Jockel, P., M. Di Bernardino, and P. Dimroth. (1999). Membrane topology of the β-subunit of the oxaloacetate decarboxylase Na+ pump from Klebsiella pneumoniae. Biochemistry 38: 13461-13472.
|
Schaffitzel, C., M. Berg, P. Dimroth, and K.M. Pos. (1998). Identification of an Na+-dependent malonate transporter of Malonomonas rubra and its dependence on two separate genes. J. Bacteriol. 180: 2689-2693.
|
Vitt, S., S. Prinz, N. Hellwig, N. Morgner, U. Ermler, and W. Buckel. (2020). Molecular and Low-Resolution Structural Characterization of the Na-Translocating Glutaconyl-CoA Decarboxylase From. Front Microbiol 11: 480.
|
Wendt, K.S., I. Schall, R. Huber, W. Buckel, and U. Jacob. (2003). Crystal structure of the carboxyltransferase subunit of the bacterial sodium ion pump glutaconyl-coenzyme A decarboxylase. EMBO J. 22: 3493-3502.
|
Woehlke, G., K. Wifling, and P. Dimroth. (1992). Sequence of the sodium ion pump oxaloacatate decarboxylase from Salmonella typhimurium. J. Biol. Chem. 267: 22798-22803.
|
| Examples: |
| TC# | Name | Organismal Type | Example |
| 3.B.1.1.1 | Na+-transporting oxalo-acetate decarboxylase. Subunit stoichiometries have been described (Balsera et al., 2011). The crystal structure of the carboxyltransferase at 1.7 A resolution shows a
dimer of alpha(8)beta(8) barrels with an active site metal ion,
identified spectroscopically as Zn2+ (Granjon et al. 2010). | Bacteria | Oxaloacetate decarboxylase of Salmonella typhimurium |
| |
| 3.B.1.1.2 | Na+-transporting methylmalonyl-CoA decarboxylase | Bacteria | Methylmalonyl-CoA decarboxylase of Veillonella parvula |
| |
| 3.B.1.1.3 | Na+-transporting glutaconyl-CoA decarboxylase | Bacteria | Glutaconyl-CoA decarboxylase of Acidaminococcus fermentans |
| |
| 3.B.1.1.4 | Na+-transporting malonate decarboxylase | Bacteria | Malonate decarboxylase of Malonomonas rubra |
| |
| 3.B.1.1.5 | Putative Na+-transporting methylmalonyl-CoA decarboxylase (Cohen et al., 2003) | Archaea | Putative methylmalonyl-CoA decarboxylase of Pyrococcus abyssi (α,β,γ,δ subunits)
α (MmdA)
β (MmdB)
γ (MmdC)
δ (MmdD) |
| |
| 3.B.1.1.6 | Na+-exporting oxaloacetate decarboxylase with three subunits, OadA (α), OadB (β) and OadG (γ) of 599 aas and 0 TMSs, 433 aas and 11 TMSs, and 90 aas and 1 TMS, respectively. These three subunits are 72%, 70% and 40% identical to 3.B.1.1.1. The stoichiometry of the three subunits is 4:2:2, and large amounts of the protein can be generated for crystalographic studies (Inoue and Li 2015). | | OadABG of Vibrio cholerae |
| |
| 3.B.1.1.7 | Na+-pumping glutaconyl-CoA decarboxylase, Gcd, with subunits A4/B2/C11/C21/D2 (The stoichiometry of the subunits is given by the subscripts.). Gcd drives the endergonic translocation of Na+ across the membrane with the exergonic decarboxylation of glutaconyl-CoA (ΔG0’ ≈−30 kJ/mol) to crotonyl-CoA. Vitt et al. 2020 reported on the molecular characterization of Gcd from Clostridium symbiosum. The subunit composition is four GcdA
(65 kDa), two GcdB (35 kDa), one GcdC1 (15 kDa), one GcdC2 (14 kDa), and two GcdD
(10 kDa). Low-resolution structural information was achieved by electron microscopic (EM) measurements, which resulted in a
3D reconstruction model based on negative-stained particles. The Gcd
structure is built up of a membrane-spanning base primarily composed of
the GcdB dimer and a solvent-exposed head with the GcdA tetramer as the
major component. These two globular parts are bridged by a linker presumably
built up of segments of GcdC1, GcdC2 and the 2 GcdDs. The structure of
the highly mobile Gcd complex represents a template for the global
architecture of the Bdc family (Vitt et al. 2020). | | Glutaconyl-CoA decarboxylase, GcdABC1C2D of Clostridium symbiosum (previously mistakenly referred to as Bacteroides symbiosus)
GcdA, 558 aas
GcdB, 278 aas and 9 TMSs
GcdC1, 149 aas and 0 - 2 TMSs
GcdC2, 139 aas and 0 - 2 TMSs
GcdD, 95 aas and 1 (or maybe 2) TMSs
|
| |
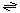 R - H CO2 1 or 2 Na+ (out)
R - H CO2 1 or 2 Na+ (out)