| 1.B.9 The FadL Outer Membrane Protein (FadL) Family
The FadL family includes several distantly related proteins, all probably outer membrane proteins, sequenced from E. coli, Haemophilus influenzae, Pseudomonas putida, Moraxella catarrhalis and Chlorobium limicola. The E. coli FadL protein functions in long chain fatty acid transport across the outer membrane. Residues involved in fatty acid binding and transport have been distinguished and identified. The XylN and TodX proteins of P. putida are encoded on TOL (toluene-degradation) plasmids and are concerned with transport of aromatic compounds such as toluene, m-xylene and benzyl alcohol. Other homologues are not characterized functionally.However a novel FadL homolog, PadL, involved in the biodegradation of the representative polycyclic aromatic hydrocarbons (PAHs) such as phenanthrene in Novosphingobium pentaromativorans US6-1 has been characterized (Meng et al. 2025). This bacterium is an efficient PAH-degrading bacterium. PadL facilitates the
cross-OM transport of phenanthrene, thus upregulating the expression of
the gene ahdA1e that is critical to PAH catabolism. Hydrophobic amino acid residues in the substrate binding
pockets of PadL are essential for the binding of PAHs, such as
phenanthrene and benzo[a]pyrene. PadL homologs commonly exist in most of the PAH-degrading species from Sphingomonas and Novosphingobium (Meng et al. 2025).
Proteins of the FadL family are of about 450 amino acyl residues in length. They exhibit a single N-terminal hydrophobic sequence that may serve as the cleavable membrane-targeting signal sequence. The remainder of the proteins exhibit a preponderance of β-structure which forms β-barrels as do other bacterial porins. However, in contrast to most structurally characterized porins, FadL is monomeric.
The 3-dimensional structure of FadL has been solved at 2.6 Å resolution (van den Berg et al., 2004). It forms a monomeric 14-stranded β-barrel that is occluded by a central hatch domain. The hydrophobic compounds probably bind to multiple sites in the FadL channel and use a transport mechanism that involves spontaneous conformational changes in the hatch region.
Hearn et al., 2009 presented an example of a lateral diffusion mechanism for the uptake of hydrophobic substrates by the Escherichia coli FadL. A FadL mutant, in which a lateral opening in the barrel wall was constricted, but otherwise unalterred, did not transport substrates. A crystal structure of FadL from Pseudomonas aeruginosa showed that the opening in the wall of the beta-barrel is conserved and delineates a long, hydrophobic tunnel that can mediate substrate passage from the extracellular environment, through the polar lipopolysaccharide layer. By means of the lateral opening in the barrel wall, substrates can pass into the lipid bilayer from where they can diffuse into the periplasm (Hearn et al., 2009).
Regular phospholipid bilayers do not pose efficient barriers for the transport of hydrophobic molecules. The outer membrane (OM) surrounding Gram-negative bacteria is a nontypical, asymmetric bilayer with an outer layer of lipopolysaccharide (LPS). The sugar molecules of the LPS layer prevent spontaneous diffusion of hydrophobic molecules across the OM. As regular OM channels such as porins do not allow passage of hydrophobic molecules, specialized OM transport proteins are required for their uptake. Such proteins, exemplified by channels of the FadL family, transport their substrates according to a lateral diffusion mechanism. Substrates diffuse from the lumen of the β-barrel laterally into the OM, through a stable opening in the wall of the barrel. In this way, the lipopolysaccharide barrier is bypassed, and, by depositing the substrates into the OM, a driving force for uptake is provided. Lateral diffusion through protein channel walls also occurs in alpha-helical inner membrane proteins, and could represent a widespread mechanism for proteins that transport and interact with hydrophobic substrates (van den Berg, 2010) .
The generalized transport reaction catalyzed by FadL family proteins is:
Hydrophobic compound (out) 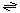 hydrophobic compound (periplasm) hydrophobic compound (periplasm)
|
This family belongs to the Outer Membrane Pore-forming Protein I (OMPP-I) Superfamily .
|
| References: |
Bhat, S., X. Zhu, R.P. Patel, R. Orlando, and L.J. Shimkets. (2011). Identification and localization of Myxococcus xanthus porins and lipoproteins. PLoS One 6: e27475.
|
Black, P.N. (1991). Primary sequence of the Escherichia coli fadL gene encoding an outer membrane protein required for long-chain fatty acid transport. J. Bacteriol. 173: 435-442.
|
Gregson, B.H., G. Metodieva, M.V. Metodiev, P.N. Golyshin, and B.A. McKew. (2018). Differential Protein Expression During Growth on Medium Versus Long-Chain Alkanes in the Obligate Marine Hydrocarbon-Degrading Bacterium MIL-1. Front Microbiol 9: 3130.
|
Hearn, E.M., D.R. Patel, and B. van den Berg. (2008). Outer-membrane transport of aromatic hydrocarbons as a first step in biodegradation. Proc. Natl. Acad. Sci. USA 105: 8601-8606.
|
Hearn, E.M., D.R. Patel, B.W. Lepore, M. Indic, and B. van den Berg. (2009). Transmembrane passage of hydrophobic compounds through a protein channel wall. Nature 458: 367-370.
|
Jones, R.M., L.S. Collier, E.L. Neidle, and P.A. Williams. (1999). areABC genes determine the catabolism of aryl esters in Acinetobacter sp. strain ADPI. J. Bacteriol. 181: 4568-4575.
|
Jones, R.M., V. Pagmantidis, and P.A. Williams. (2000). sal genes determining the catabolism of salicylate esters are part of a supraoperonic cluster of catabolic genes in Acinetobacter sp. strain ADP1. J. Bacteriol. 182: 2018-2025.
|
Kasai, Y., J. Inoue, and S. Harayama. (2001). The TOL plasmid pWWO xylN gene product from Pseudomonas putida is involved in m-xylene uptake. J. Bacteriol. 183: 6662-6666.
|
Kumar, G.B. and P.N. Black. (1993). Bacterial long-chain fatty acid transport. Identification of amino acid residues within the outer membrane protein FadL required for activity. J. Biol. Chem. 268: 15469-15476.
|
Lepore, B.W., M. Indic, H. Pham, E.M. Hearn, D.R. Patel, and B. van den Berg. (2011). Ligand-gated diffusion across the bacterial outer membrane. Proc. Natl. Acad. Sci. USA 108: 10121-10126.
|
Meng, Q., Y. Liang, Y. Xu, S. Li, H. Huang, Y. Xu, F. Cao, J. Yin, T. Zhu, H. Gao, and Z. Yu. (2025). A novel FadL family outer membrane transporter is involved in the uptake of polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 91: e0082724.
|
van den Berg, B. (2010). Going forward laterally: transmembrane passage of hydrophobic molecules through protein channel walls. Chembiochem 11: 1339-1343.
|
van den Berg, B., P.N. Black, W.M. Clemons, Jr., and T.A. Rapoport. (2004). Crystal structure of the long-chain fatty acid transporter FadL. Science 304: 1506-1509.
|
Verardi, R., L. Shi, N.J. Traaseth, N. Walsh, and G. Veglia. (2011). Structural topology of phospholamban pentamer in lipid bilayers by a hybrid solution and solid-state NMR method. Proc. Natl. Acad. Sci. USA 108: 9101-9106.
|
Wang, Y., M. Rawlings, D.T. Gibson, D. Labbe, H. Bergeron, R. Brousseau, and P.C. Lau. (1995). Identification of a membrane protein and a truncated LysR-type regulator associated with the toluene degradation pathway in Pseudomonas putida Fl. Mol. Gen. Genet. 246: 570-579.
|
Yuan, M.Q., Z.Y. Shi, X.X. Wei, Q. Wu, S.F. Chen, and G.Q. Chen. (2008). Microbial production of medium-chain-length 3-hydroxyalkanoic acids by recombinant Pseudomonas putida KT2442 harboring genes fadL, fadD and phaZ. FEMS Microbiol. Lett. 283: 167-175.
|
Zhang, H., H. Zhang, B. Xiong, G. Fan, and Z. Cao. (2019). Immunogenicity of recombinant outer membrane porin protein and protective efficacy against lethal challenge with Bordetella bronchiseptica in rabbits. J Appl Microbiol 127: 1646-1655.
|
Zhou J., Wang K., Xu S., Wu J., Liu P., Du G., Li J. and Chen J. (2015). Identification of membrane proteins associated with phenylpropanoid tolerance and transport in Escherichia coli BL21. J Proteomics. 113:15-28.
|
| Examples: |
| TC# | Name | Organismal Type | Example |
| 1.B.9.1.1 | Fatty acid outer membrane porin. Gated by high affinity ligand (fatty acid) binding which causes conformational changes in the N-terminus that open up a channel for substrate diffusion (Lepore et al., 2011). May function in the transport of phenylpropanoids (resveratrol, naringenin and rutin) (Zhou et al. 2014). | Gram-negative bacteria | FadL of E. coli |
| |
| 1.B.9.1.2 | FadL homologue (Bhat et al. 2011). | Proteobacteria | FadL homologue of Myxococcus xanthus |
| |
| 1.B.9.1.3 | Putative porin of 441 aas | Nitrospira | Porin of Candidatus Nitrospira defluvii |
| |
| 1.B.9.1.4 | Outer membrane protein P1 of 459 aas | Proteobacteria | OmpP1 of Haemophilus influenzae |
| |
| 1.B.9.1.5 | Putative fatty acid-transporting porin of 434 aas, OmpP1, FadL, TodX. It has used to generate an effective vaccine against Bordetella bronchiseptica (Zhang et al. 2019). | | OmpP1 of Bordetella bronchiseptica (Alcaligenes bronchisepticus) |
| |
| 1.B.9.1.6 | Long alkane hydrocarbon chain (~C28) transporting outer membrane porin, FadL of 546 aas (Gregson et al. 2018). | | FadL porin of the obligate marine hydrocarbon-degrading bacterium, Thalassolituus oleivorans MIL-1. |
| |
| Examples: |
| TC# | Name | Organismal Type | Example |
| 1.B.9.2.1 | Toluene/m-xylene outer membrane porin, XylN or FadL. May also transport medium-chain-length 3-hydroxyalkanoic acids (Yuan et al. 2008). | Gram-negative bacteria | XylN of Pseudomonas putida |
| |
| 1.B.9.2.2 | The 14 TMS hydrocarbon porin, TodX. The x-ray structure is known (3BS0-A) (Hearn et al., 2008). | Bacteria | TodX of Pseudomonas putida (3BS0_A) |
| |
| 1.B.9.2.3 | The 14 TMS hydrocarbon porin, TbuX. The crystal structure is known. (3BRY_A) (Hearn et al., 2008). | Bacteria | TbuX of Ralstonia pickettii (3BRY_A)(Q9RBW8) |
| |
| 1.B.9.2.4 | Putative aromatic hydrocarbon degradation pathway porin, FadL homologue, with an N-terminal transmembrane α-helix and about 16 putative β-TMSs. | Spirochaetes | FadL homologue of Treponema succinifaciens |
| |
| Examples: |
| TC# | Name | Organismal Type | Example |
| 1.B.9.3.1 | Salicylate ester (methyl and ethyl salicylates)/hydrocarbon outer membrane porin, SalD (Jones et al. 2000). | Gram-negative bacteria | SalD of Acinetobacter sp. strain ADPI |
| |
| 1.B.9.3.2 | FadL homologue of 432 aas | Chlamydiae | FadL of Parachlamydia acanthamoebae |
| |
| 1.B.9.3.3 | FadL homologue of 468 aas | Spirochaetes | FadL homologue of Leptospira interrogans |
| |
| Examples: |
| TC# | Name | Organismal Type | Example |
| 1.B.9.4.1 | Putative hemin receptor of 479 aas and ~20 β-strand | Proteobacteria | Hemin receptor of Riemerella anatipestifer |
| |
| 1.B.9.4.2 | Uncharacterized porin of 542 aas | Proteobacteria | UP of Prevotella oralis |
| |
| 1.B.9.4.3 | Putative porin of 543 aas | Bacteroidetes | Porin of Porphyromonas endodontalis |
| |
| 1.B.9.4.4 | Outer membrane protein transport protein (OmpP1/FadL/TodX family) | Bacteroidetes | OmpP1 of Saprospira grandis |
| |
| 1.B.9.4.5 | Membrane protein involved in aromatic hydrocarbon degradation of 481 aas | Caldithrix | OM porin of Caldithrix abyssi |
| |
| 1.B.9.4.6 | Uncharacterized protein of 472 aas | Ignavibacteriae | UP of Ignavibacterium album |
| |
| 1.B.9.4.7 | Porin protein involved in aromatic hydrocarbon degradation; putative hemin receptor of 479 aas
| Bacteroidetes | Putative porin of Pedobacter heparinus |
| |
| 1.B.9.4.8 | Uncharacterized membrane protein, predicted to be involved in aromatic hydrocarbon degradation; of 437 aas. | Planctomycetes | UP of planctomycete KSU-1 |
| |
 hydrophobic compound (periplasm)
hydrophobic compound (periplasm)