9.A.32 The SdpC (Peptide-Antibiotic Killer Factor) Immunity Protein, SdpI (SdpI) Family
SdpI (YvaZ) of Bacillus subtilis (COG5658) provides immunity to a peptide antibiotic killer factor, SdpC, possibly by exporting the toxic factor (Ellermeier et al., 2006). It is a 6 TMS protein of about 200 residues with homologues of similar size in a wide variety of bacterial and archaeal kingdoms. However, some homologues are larger (e.g., Deinococcus geothermalis with 12 TMSs and 404 aas) (a probable duplication, based on the hydropathy profile), while others are smaller or larger, with 3 or 4 putative TMSs. Several are annotated as putative 'tryptophan permease' (e.g., gi25029421, 280 aas and gi629190, 170 aas, both of C. glutamicum, both with 3 putative TMSs.
SdpI family members are transmembrane proteins with 3, 4, 5, 6, 7, 8, or 12 putative TMSs. These varied topologies appear to be genuine rather than artifacts due to sequencing or annotation errors. The basic and most frequently occurring element of the SdpI family has 6 TMSs. Homologues of all topological types have been aligned to determine the homologous TMSs and loop regions, and the positive-inside rule was used to determine sidedness (Povolotsky et al. 2010). The two most conserved motifs were identified between TMSs 1 and 2 and TMSs 4 and 5 of the 6 TMS proteins. These showed significant sequence similarity, leading to the suggestion that the primordial precursor of these proteins was a 3 TMS-encoding genetic element that underwent intragenic duplication. Various deletional and fusional events, as well as intragenic duplications and inversions, may have yielded SdpI homologues with topologies of varying numbers and positions of TMSs. Povolotsky et al. (2010) proposed a specific evolutionary pathway that could have given rise to these distantly related bacterial immunity proteins. Genes encoding SdpI homologues often appear in operons with genes for homologues of SdpR, SdpI's autorepressor. Structure-function relationships were proposed that may be applicable to most family members.
Several of these proteins showed alignments with SdpI that indicate that transposition of one protein segment relative to the other had occurred. For instance, after the 2nd iteration, homologues from both Archaeoglobus fulgidus (gi11499784; 228 aas; 6 TMSs) and Deinococcus geothermalis (gi66797038; 404 aas; 12 TMSs) were retrieved that showed sequence similarity: Residues 110-204 of SdpI aligned with residues 6-103 of the A. fulgidus homologue, and residues 22-69 aligned with residues 141-190 of the A. fulgidus protein. Further, residues 105-179 of SdpI aligned with residues 87-161 of D. geothermalis, and residues 20-62 of SdpI aligned with residues 232-274 of D. geothermalis (Povolotsky et al. 2010). None of these sequences exhibit significant similarity with anything else in TCDB.
The presumed reaction catalyzed by SdpI of B. subtilis is:
peptide (in) 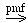 peptide (out)
peptide (out)
That for the putative tryptophan (methyltryptophan resistance protein) is:
tryptophan (in) 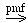 tryptophan (out)
tryptophan (out)