9.A.36 The Ca2+-dependent Phospholipid Scramblase (Scramblase) Family
Phospholipid scramblases are a group of homologous proteins that are conserved in all eukaryotic and some prokaryotic organisms. They are believed to be involved in destroying plasma membrane phospholipid asymmetry at critical cellular events like cell activation, injury and apoptosis. However, a detailed mechanism of phospholipid scrambling still awaits elucidation. The most studied member of this family, phospholipid scramblase 1 (PLSCR1) (a 37 kDa protein), is involved in rapid Ca2+-dependent transbilayer redistribution of plasma membrane phospholipids (Pomorski and Menon, 2006; Sahu et al., 2007). The function of PLSCR1 as a phospholipid translocator has been challenged and that PLSCR1 acts as signaling molecule. It has been shown to be involved in protein phosphorylation and as a potential activator of genes in response to interferon and other cytokines. Interferon induced rapid biosynthesis of PLSCR1, targets some of the protein into the nucleus, where it binds to the promoter region of the inositol 1,4,5-triphosphate (IP3) receptor type 1 (IP3R1) gene and induces its expression. Palmitoylation of PLSCR1 acts as a switch, controlling its localization either to the PM or inside the nucleus (Sahu et al., 2007).
Human phospholipid scramblase 1 (SCR) is believed to be an intrinsic membrane protein catalyzing transbilayer phospholipid transfer in the absence of ATP, but a role as a nuclear transcription factor has also been proposed, either in addition or alternatively to its capacity to facilitate phospholipid flip-flop. A predicted α-helix (aa residues 288-306) located near the C-terminus has been alternatively proposed as a transmembrane domain or as a protein core structural element. Posada et al. 2013 showed that the 288-306 peptide of SCR becomes membrane inserted in the presence of lipid bilayers in agreement with the possibility that SCR is an integral membrane protein.
Human SCR consists of a large cytoplasmic domain and a small presumed transmembrane domain near the C-terminal end of the protein. A SCRΔ mutant lacking the C-terminal portion (last 28 aa) revealed the importance of this C-terminal moiety for protein function and calcium-binding affinity. The cytoplasmic domain showed high affinity for lipid membranes and behaved like an intrinsic membrane protein. Phosphatidylserine may be important for the SCR cytoplasmic domain to be electrostatically anchored to the plasma membrane inner surface. SCR is an integral membrane protein in which both the transmembrane domain and the cytoplasmic moiety play a role in membrane docking.
Scramblases have properties that facilitate lipid flip-flop from one membrane leaflet to another. Scramblases and similar transmembrane proteins could also affect the translocation of other amphiphilic molecules, including cell-penetrating (CP) or antimicrobial peptides (AMPs). Bartoš et al. 2021 studied the effect of transmembrane proteins on the translocation of amphiphilic peptides through the membrane. They demonstrated that transmembrane proteins with a hydrophilic patch enhance the translocation of amphiphilic peptides by stabilizing the peptide in the membrane.
Lipid transbilayer movement (flip-flop) is regulated by membrane proteins that are involved in homeostasis and signaling in eukaryotic cells. In the plasma membrane, an asymmetric lipid composition is maintained by energy-dependent unidirectional transport. Energy-independent flip-flop promotion by phospholipid scramblases disrupts the asymmetry in several physiological processes, such as apoptosis and blood coagulation. In the endoplasmic reticulum, rapid flip-flop is essential for bilayer integrity because phospholipids are synthesized only in the cytoplasmic leaflet. Phospholipid scramblases are also involved in lipoprotein biogenesis, autophagosome formation, and viral infection. Although several scramblases have been identified and investigated, the precise flip-flop promotion mechanisms are not fully understood. Model transmembrane peptides are valuable tools for investigating the general effects of lipid-peptide interactions (Nakao and Nakano 2022).
The generalized transport reaction proposed for scramblases is:
phospholipid (inner leaflet) 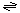 phospholipid (outer leaflet)
phospholipid (outer leaflet)